Abstract
Background: The prognostic importance of 1q gain, present in 30% of cases, has led to it being extensively studied and incorporated into prognostic systems. Amplification (>3 copies) of 1q is seen in 10% of cases and has a greater degree of specificity for poor outcome than gain alone. Despite this, there is ongoing debate about its value, likely due to differences in the nature and penetrance of deregulated drivers. To gain insight into this we dissected the structural changes seen at the 1q level and integrated them with expression and clinical data to identify potential drivers and gain insight into progression mechanisms.
Methods We analyzed data derived from 1,154 CoMMpass trial patients including 972 NDMM patients with whole exome for mutations, and 752 whole genomes for copy number (CNA), translocations, and complex rearrangements. Using GISTIC 2.0 and PFC we identified hotspots of CNA. This information was analyzed in conjunction with the RNA-seq data derived from 643 patients to determine the aberrant transcriptional landscape of 1q.
Results: We identified nine regions of gain (numbered G1 to G9), eight of them being on 1q. Several potential oncogenes such as BCL9 (G2), MCL1 (G3), SLAMF7 (G5), POU2F1 (G6), and BTG2 (G8) lay within these regions of gain. Of note, other previously identified drivers lay outside specific hotspots: ANP32E (306 kb downstream of G3) CKS1B and ADAR1 (1209 kb ( and 1561 kb away from G4, respectively), ATF6 (941 kb from G5), and PBX1 (2313 kb upstream of G6). These data suggest that although important, these hotpots do not drive the entire spectrum of genes upregulated as a consequence of gain 1q.
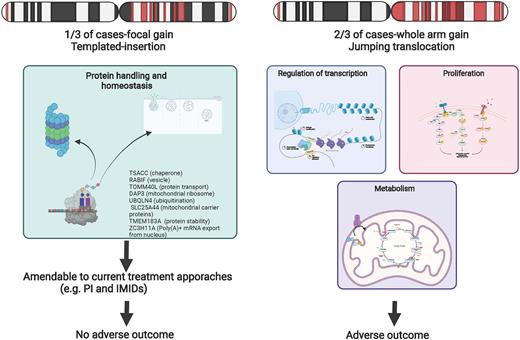
We noted that 302 samples (40%) had at least one gained region with 80% (241/302) having gain of 7-9 regions; 69% (207/302) had gain G2-G9. Each of the gained regions occurred at a similar frequency (G2-G9 range 79-87%) with the exception of G1 located at the sub-telomeric border of 1p seen only in 6% (18/302) of patients. There were 219 cases with gain of G2-G9. These results show two broad groups of gain 1q; whole arm gains and a group with focal gain.
Patients with whole arm gain had a worse PFS (HR 1.3 (95% CI 1.01-1.6) and OS (HR 1.6 (95% CI 1.15-2.2) than patients with no 1q gain; the outcome for patients focal events was neutral. Whole arm gains strongly associated with a complex genomic background including with t(4;14) (corr=0.10, BF=1.06), loss of acrocentric chromosomes (del(13q) corr=0.23, BF=118623 and del(14q) corr=0.12, BF=2.9) and other deletions (del(4p) corr=0.11, BF=1.89, del(16p) corr=0.12, BF=2.3) and negatively correlated to trisomies and t(11;14) (corr=0.16, BF=184). Interestingly whole arm gains had significantly shorter telomeres and a higher mean age, and patients with critically short tumor telomeres were over-represented (Corr=0.11, BF=1.2). Common associations with CKS1B gain are mostly found in the whole arm gains group. Amp(1q) was over represented in the whole arm gain group (87%) and associated with the worst outcome (data not shown). Similarly, focal events strongly associated with templated insertion (corr=0.34, BF=6e14), MYC translocation (corr=0.24, BF=3034538), and HRD (corr=0.13, BF=4.9).
To address expression changes in focal changes we compared patients with no 1q gain (n=297) with patients with a focal event (n=56); we identified 20 genes of which the majority (n=12) were located on chromosome 1 and related to protein handling. We compared patients with no evidence of 1q gain with patients with a whole arm gain (n= 157); the number of deregulated genes was higher both on 1q and overall (n=585 and n=2409 respectively) suggesting that generalized gene deregulation is part of a more complex genome wide mechanism. Interestingly, 101 were transcription factors and included upregulation of BACH2, NR5A1, MYBL1. GLMP, and E2F2 and downregulation of PAX5, SMAD1, TBX21, SIX4, EGR3, and GF1 most of them being involved in histone methylation, intracellular signaling, and regulation of transcription.
Conclusions: Our data add precision by which 1q is associated with an adverse outcome and an aggressive disease phenotype by separating out focal gains, that occur on a favorable background and associate with a good outcome and those with arm-length gains, associated with high-risk disease features, deregulation of multiple transcriptional programs, increased metabolism and aggressive clinical behavior.
Disclosures
Braunstein:Abbvie: Membership on an entity's Board of Directors or advisory committees; ADC Therapeutics: Membership on an entity's Board of Directors or advisory committees; Amgen: Membership on an entity's Board of Directors or advisory committees; BMS: Membership on an entity's Board of Directors or advisory committees; Epizyme: Membership on an entity's Board of Directors or advisory committees; GSK: Membership on an entity's Board of Directors or advisory committees; Janssen: Membership on an entity's Board of Directors or advisory committees; Morphosys: Membership on an entity's Board of Directors or advisory committees; Pfizer: Membership on an entity's Board of Directors or advisory committees; TG Therapeutics: Membership on an entity's Board of Directors or advisory committees. Landgren:Aptitude Health: Honoraria; MMRF: Honoraria; Leukemia & Lymphoma Society: Research Funding; Rising Tide Foundation: Research Funding; Theradex: Other: Independent Data Monitoring Committee (IDMC) member for clinical trials; NCI/NIH: Research Funding; Riney Foundation: Research Funding; Tow Foundation: Research Funding; Merck & Co., Inc.: Other: Independent Data Monitoring Committee (IDMC) member for clinical trials; Janssen: Honoraria, Other: Independent Data Monitoring Committee (IDMC) member for clinical trials, Research Funding; Pfizer Inc: Consultancy; Amgen: Honoraria, Research Funding. Walker:Genentech: Research Funding; Bristol Myers Squibb: Research Funding. Davies:Janssen: Consultancy, Honoraria; Celgene/BMS: Consultancy, Honoraria; Roche: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; Abbvie: Consultancy, Honoraria; Takeda: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal